4. EOS Storage
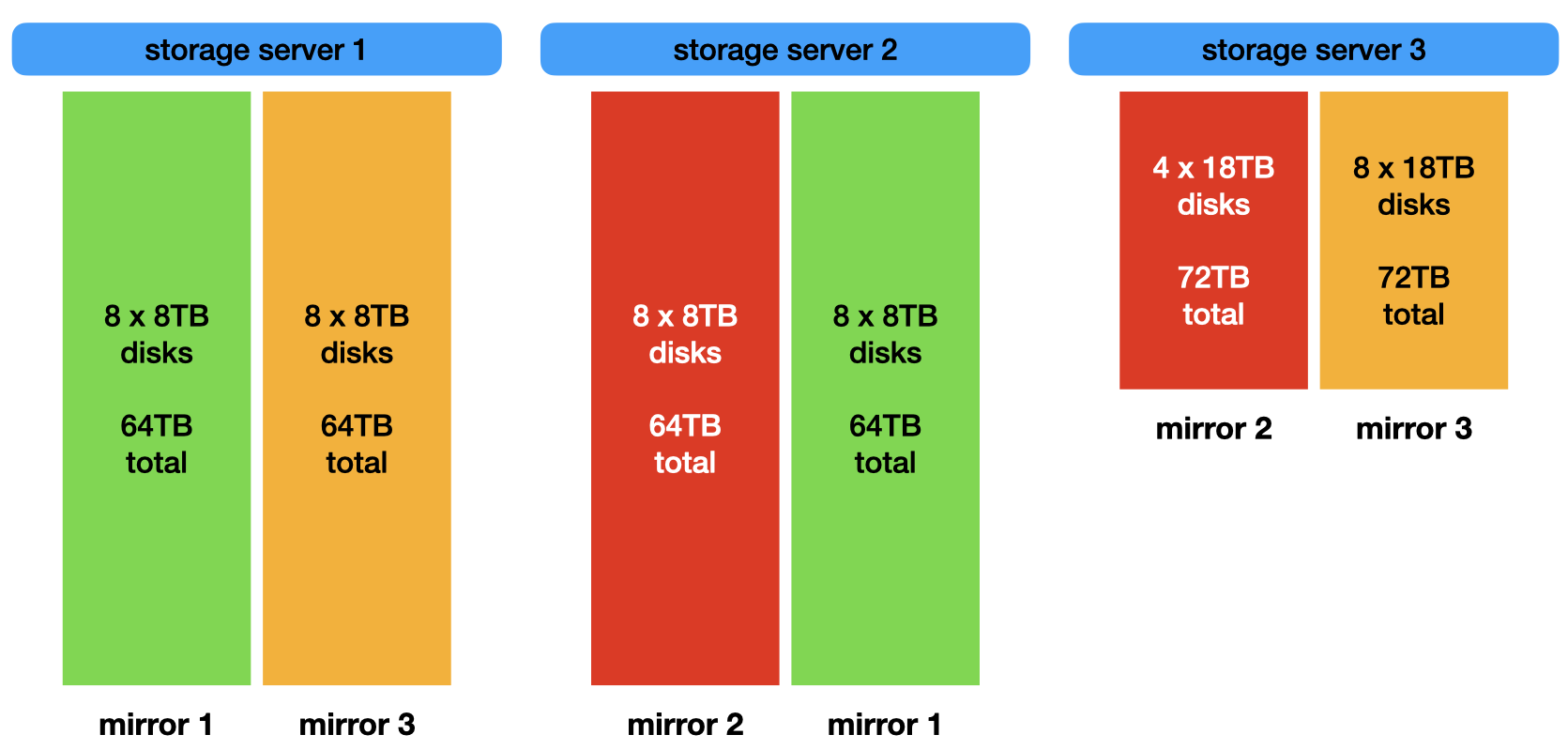
4.1. Storage Configuration
4.2. Large files and common resources
Warning
Strict guidelines should be followed for downloading large files like databases, references, protein structure data and the likes.
These files are very likely to be of common interest to other users, and we need to avoid multiplication of these resources in each user’s home, thus reducing the total storage available for files that could be simply shared.
These files should be placed in a special folder:
/home/shared
This folder is special because it is not mirrored on disk (i.e. placed in the highest tier of data safety). The folder is however in stripe, i.e. on RAID0 which ensures a certain level of redundancy and recovery of the data, but does not occupy double the disk space like all other data placed in /home. RAID0 also ensures higher speed reads and writes, and it is therefore most suitable for highly accessed and higher volume data like these.
Note
Since these data are going to be shared among all users, it is important that:
before adding new data, please check whether the data already exist within this location
data are categorised according to very general domains, which might be arbitrary: please follow existing structure if the data you are interested into falls already under another category in this catalogue
data can be added, but should NOT be deleted unless agreed with the HPC governance team
In order to keep track of the data added to this folder, a README file should be maintained with everyone’s contribution.
The file is located at:
/home/shared/README
Please update this file and follow the guidelines when adding new data
4.3. Genomic references
Warning
Please, maintain the current structure for these data which follows the de-facto standard of Illumina iGenomes repository.
In order to add new data to this folder, we commend using the aws-cli and downloading the data with the command you can build using this tool:
This allows you to download data with a command like this (for example for Mus Musculus):
aws s3 --no-sign-request --region eu-west-1 sync s3://ngi-igenomes/igenomes/Mus_musculus/Ensembl/GRCm38 ./references/Mus_musculus/Ensembl/GRCm38
which preserves the existing structure in the folder.